fragment_assembly (RELEASE_2.5.1) |
|
|---|---|
| SO Accession: | SO:0001249 (SOWiki) |
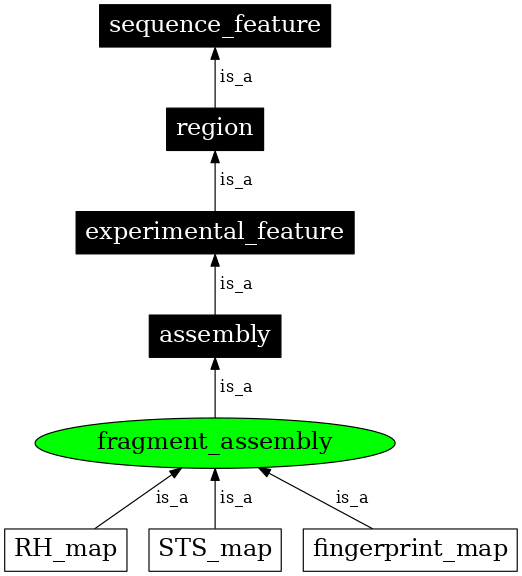
| Definition: | A fragment assembly is a genome assembly that orders overlapping fragments of the genome based on landmark sequences. The base pair distance between the landmarks is known allowing additivity of lengths. |
| Synonyms: | fragment assembly, physical map |
| DB Xrefs: | SO: ke |
| Parent: | assembly (SO:0001248) |
| Children: | fingerprint_map (SO:0001250) |
| STS_map (SO:0001251) | |
| RH_map (SO:0001252) | |
In the image below graph nodes link to the appropriate terms. Clicking the image background will toggle the image between large and small formats.