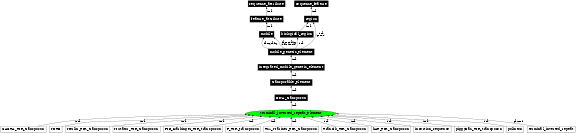
terminal_inverted_repeat_element (CURRENT_SVN) |
|
|---|---|
| SO Accession: | SO:0000208 (SOWiki) |
| Definition: | A DNA transposable element defined as having termini with perfect, or nearly perfect short inverted repeats, generally 10 - 40 nucleotides long. |
| Synonyms: | terminal inverted repeat element, TIR element |
| DB Xrefs: | URL: http://www.genetics.org/cgi/reprint/156/4/1983.pdf |
| Parent: | DNA_transposon (SO:0000182) |
| Children: | Transib_TIR_transposon (SO:0002282) |
| Merlin_TIR_transposon (SO:0002281) | |
| piggyBac_TIR_transposon (SO:0002283) | |
| Mutator_TIR_transposon (SO:0002280) | |
| polinton (SO:0001170) | |
| CACTA_TIR_transposon (SO:0002285) | |
| P_TIR_transposon (SO:0001535) | |
| terminal_inverted_repeat (SO:0000481) | |
| Tc1_Mariner_TIR_transposon (SO:0002278) | |
| PIF_Harbinger_TIR_transposon (SO:0002284) | |
| hAT_TIR_transposon (SO:0002279) | |
| insertion_sequence (SO:0000973) | |
| MITE (SO:0000338) | |
In the image below graph nodes link to the appropriate terms. Clicking the image background will toggle the image between large and small formats.