DsrA_RNA (CURRENT_RELEASE) |
|
|---|---|
| SO Accession: | SO:0000378 (SOWiki) |
| Definition: | DsrA RNA regulates both transcription, by overcoming transcriptional silencing by the nucleoid-associated H-NS protein, and translation, by promoting efficient translation of the stress sigma factor, RpoS. These two activities of DsrA can be separated by mutation: the first of three stem-loops of the 85 nucleotide RNA is necessary for RpoS translation but not for anti-H-NS action, while the second stem-loop is essential for antisilencing and less critical for RpoS translation. The third stem-loop, which behaves as a transcription terminator, can be substituted by the trp transcription terminator without loss of either DsrA function. The sequence of the first stem-loop of DsrA is complementary with the upstream leader portion of RpoS messenger RNA, suggesting that pairing of DsrA with the RpoS message might be important for translational regulation. |
| Synonyms: | DsrA RNA |
| DB Xrefs: | URL: http://www.sanger.ac.uk/cgi-bin/Rfam/getacc?RF00014 |
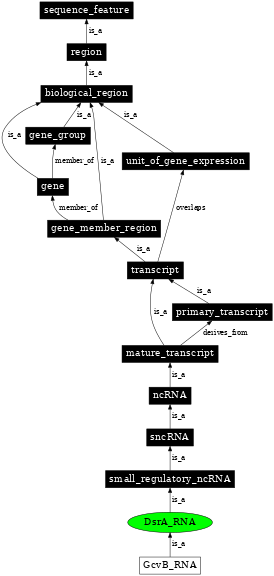
| Parent: | small_regulatory_ncRNA (SO:0000370) |
| Child: | GcvB_RNA (SO:0000379) |
In the image below graph nodes link to the appropriate terms. Clicking the image background will toggle the image between large and small formats.